Article in J. Am. Soc. Mass Spectrom
Published:
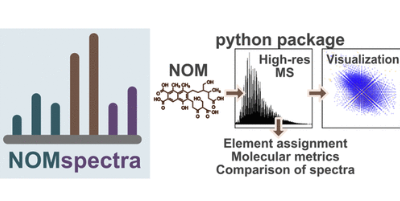
I am happy to share my latest published work, which has turned out to be not typical but holds very significance for me, and I hope for others too. In this work, I didn’t conduct any experiments in the laboratory. Instead, I relied solely on my laptop. My intention to develop programming skills originated from a hobby I pursued six years ago, where I created an electronic marking system for sport orienting Sportiduino. A community emerged around the project and continues to thrive and evolve, yet without me. Subsequently, I began applying my skills directly to my laboratory work, focusing on data processing and visualization. In my previous lab, strong projects emphasized mass spectrometry of complex objects - humic substances. Scripts and a Python library prototype for spectral analysis were created to solve these tasks. A year ago, I completed refining and publishing this project, resulting in a highly engaging and rewarding experience. I wrote numerous lines of code, prepared documentation, implemented automated tests, and developed a graphical interface. While professional programmers may find a lot of areas that require refinement and optimization (which I welcome through issue submissions), I am delighted with the outcome anyway. The project is open, and I am pleased to share it:
Abstract
The present study introduces NOMspectra, a Python package for processing high resolution mass spectrometry data on complex systems of natural organic matter (NOM). NOM is characterized by multicomponent composition reflected as thousands of signals producing very complex patterns in high resolution mass spectra. This complexity sets special demands on the methods of data processing used for analysis. The developed NOMspectra package offers a comprehensive workflow for processing, analyzing, and visualizing information-rich mass spectra of NOM and HS including algorithms for filtering spectra, recalibrating, and assigning elemental compositions to molecular ions. Additionally, the package includes functions for calculating various molecular descriptors and methods for data visualization. A graphical user interface (GUI) has been developed to make a user-friendly interface for the proposed package